The eDNA Consortium provides BP Sentry (formerly BulletProof) a Continuous Model Probabilistic Genotyping and Mixture Deconvolution software.
BP Sentry includes a suite of tools commonly used in a forensic DNA and / or Human Identification laboratory.
BP Sentry includes several seamless robust tools:
- Probabilistic Genotyping
- Deconvolutions
- Database searching based on PG methodologies
- BP Brutus for complex kinship and unidentified human remains
Request full access to BP Sentry
Forensic Laboratories in the United States are slow in moving towards Probabilistic Genotyping: Why?
- The “Commercial” software is too expensive…
- Join the eDNA Consortium and take advantage of the many free calculation tools
- The dread of validating a new calculation procedure
- The eDNA Consortium has developed a comprehensive User Guide, training program, and companion Manual Verification Spreadsheet doing much of the validation work for you.
- The fear of overly complex calculations—and the ability to adequately testify to the results
- The eDNA Consortium is cognizant of the “Black Box Fear” when implementing software solutions—Therefore BP Sentry tools include a comprehensive Step by Step calculation manual, numerous Published Papers, and multiple Spreadsheet Solutions demonstrating the PG algorithm in action—and full concordance.
- Become comfortable using BP Sentry with onsite PG training provided by recognized experts in the field.
- Join the eDNA Probabilistic Genotyping User’s group for sharing information among colleagues.
- The perception that calculations can take hours or days to perform
- Unlike Commercial software solutions, BP Sentry performs calculations in seconds to minutes.
- The perception that results are not reproducible
- Unlike some MCMC software solutions, BP Sentry produces consistent results each time the same scenario is calculated. When using BP Sentry the expert will never need to face the question, “Is it true your results are not reproducible?”
Why is Probabilistic Genotyping necessary.
The trend in the type of samples Forensic Laboratories are asked to resolve, along with improved extraction methods, amplification chemistries, and improved CE instrumentation have exceeded the ability to adequately resolve mixtures using binary mixture interpretation protocols. When laboratories try to analyze highly complex mixtures, such as “touch” items with multiple contributors and stochastic data, binary methods (CPE, CPI, Modified RMP) fail miserably. With traditional binary methods there exists no way to factor uncertainty.
Current strategies to evaluate low-level mixtures with dropout using the binary methods are insufficient. The existing CPE / CPI methods have no fundamental validity, there exists no unambiguous and rigorous mathematical proofs—it is fundamentally flawed and at best, a “stab” at the weight of the evidence.
What is Probabilistic Genotyping?
Years ago, the ISFG published recommendations for the interpretation of low-level mixtures when dropout is possible (Gill et al. 2012). eDNA’s Probabilistic Genotyping tools provides the ability to analyze complex mixtures by accounting for “uncertainty” in the LR.
Quoting the 2016 SWGDAM Guidelines for Validation of Probabilistic Genotyping Systems:
“Probabilistic genotyping refers to the use of biological modeling, statistical theory, computer algorithms, and probability distributions to calculate likelihood ratios (LRs) and/or infer genotypes for the DNA typing results of forensic samples (“forensic DNA typing results”). Human interpretation and review is required for the interpretation of forensic DNA typing results in accordance with the FBI Director’s Quality Assurance Standards for Forensic DNA Testing Laboratories . Probabilistic genotyping is a tool to assist the DNA analyst in the interpretation of forensic DNA typing results. Probabilistic genotyping is not intended to replace the human evaluation of the forensic DNA typing results or the human review of the output prior to reporting.”
BP Sentry
BP Sentry provides a high level of automation. The Maximum Likelihood approach is based on the frequentist inference maximizing the likelihood function with respect to the unknown parameters to obtain the maximum likelihood estimate.
The software optimizes the Likelihood (under each hypothesis) as a function of the unknown parameters in the continuous model:
1) Mix-Proportion: (mx1,…, mxC): mixture proportion for contributor 1,..,C.
2) PH Expectation: mean of a heterozygote peak height allele
3) PH Variability: coefficient of variance for a heterozygote peak height allele
4) Degradation: degradation slope
5) Stutter Prop: (n-1) and (n + 1) stutter proportions
Laboratory, Equipment, and PCR Kit Agnostic!
The exact method is used for estimating genotypes in conjunction with three unique models to determine the parameters which provide the best fit models. For these reasons BP Sentry does not require laboratory specific “training sets of data”, overly complex validation, and limitations on use of equipment or PCR amplification chemistries.
This also makes inter and intra-lab data simpler to manage when running multiple chemistries on multiple pieces of equipment. E.g. 3500 vs. 3500XL & Fusion 6C vs. GlobalFiler.
The BP Sentry Run Wizard makes it simple to Setup and Queue a Probabilistic Genotyping calculation runs.
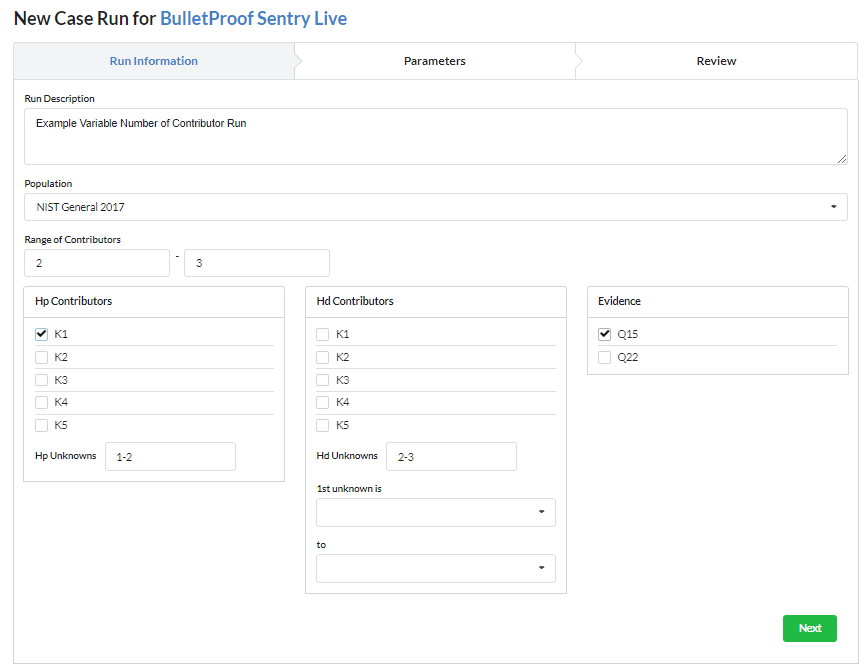
All new runs are populated with the organization’s default run parameters but can be changed based on the sample specifics.
In this example we will ask BulletProof to determine if two or three contributors best explains the evidence.
Multiple Users can simultaneously Queue runs, eliminating the expense for multiple software licenses. BulletProof supports multiple simultaneous users thus removing PG as a laboratory workflow bottleneck.
During the six year development of BP Sentry we incorporated improved workflows stemming from “lessons learned” and user feedback of other PG software tools.
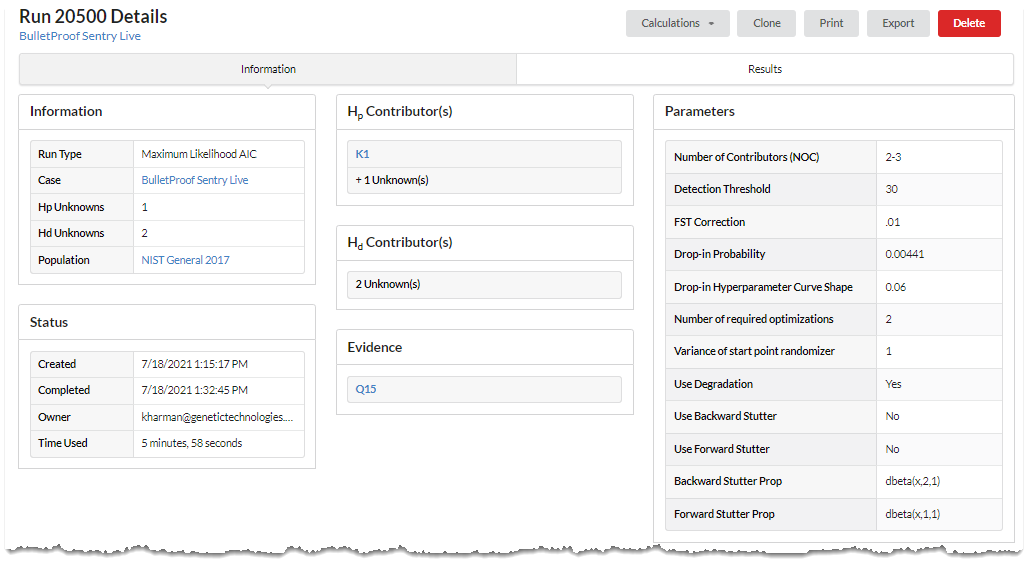
In the time it required to compose the past few sentences, BP Sentry completed the example run (detailed above). BP Sentry determined the best fit number of contributors.
Request full access to begin using these tools today.
By selecting the Results tab the user will explore a vast array of intuitive yet comprehensive PG run details.
Notice the weight of evidence is calculated using three methods; Maximum Likelihood, Bayesian Numerical, and Bayesian MCMC (Sensitivity is reported as the 5% quantile of Bayesian MCMC). Each model is based on a different mathematical algorithm used to estimate the parameters best explaining the evidence.
The Non-contributor analysis substitutes hundreds of known Non-contributors for the Person of Interest to measure the strength of the reported Likelihood ratios.
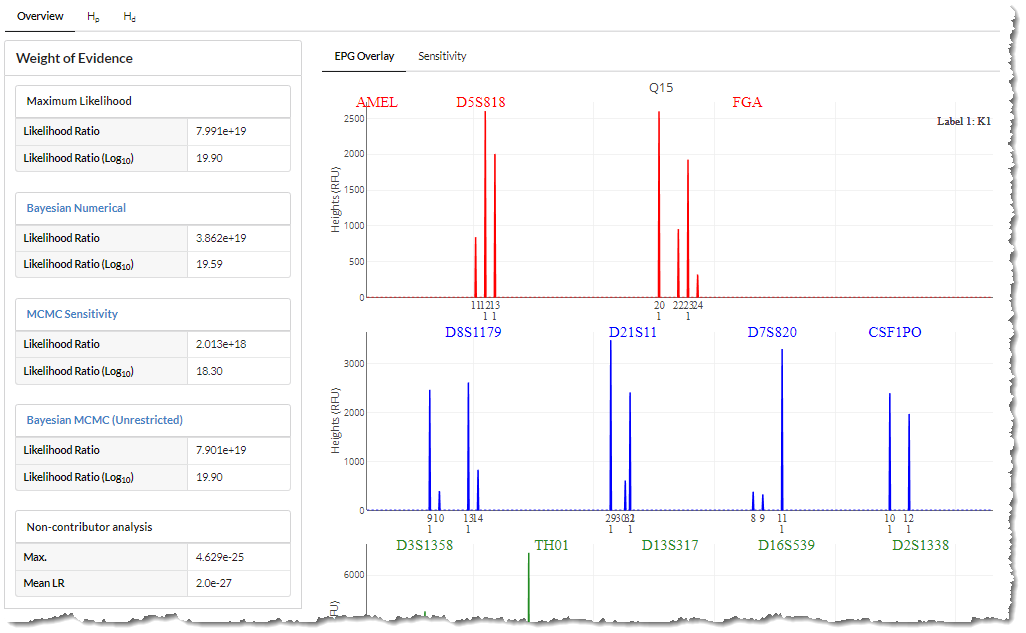
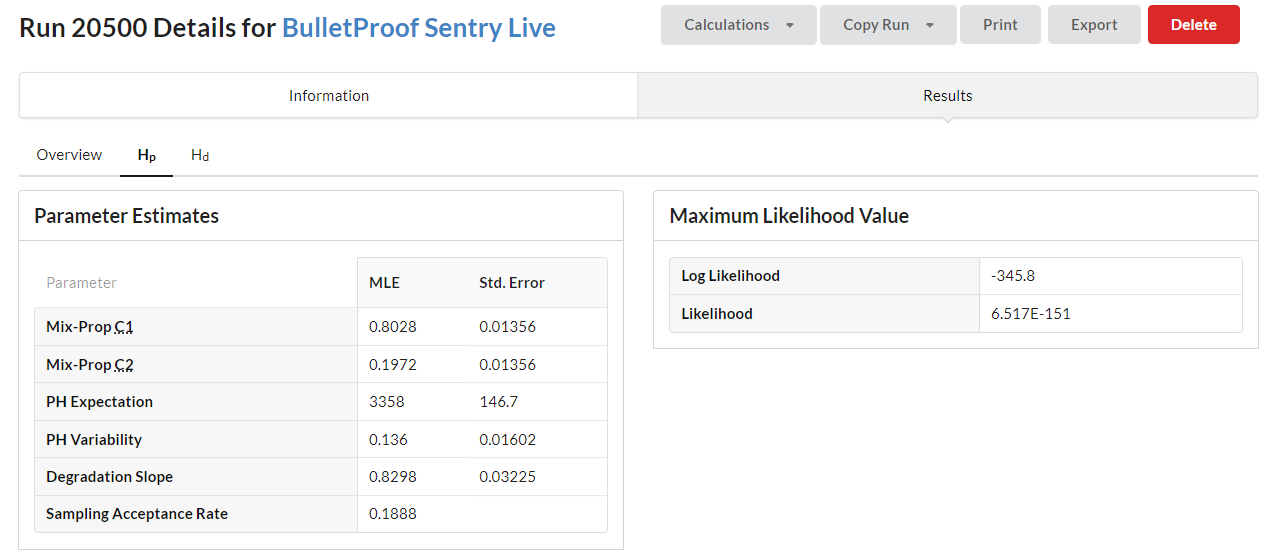
The user can further explore the results by selecting the Hp and Hd tabs.
The Hp tab below shows the mixture proportions of the two contributors with the Peak Height Expectation and Variability used in the gamma distribution.
The Hp tab also includes graphs providing a visual cue as to how well BP Sentry modeled the data. The PP-Plot is a quick way to assess the expected vs. observed probabilities.
Pages: 1 2 3 4 5
